A. Anti-viral drugs
A. Development of anti-viral drugs
Novel Dengue virus NS2B/NS3 protease inhibitors
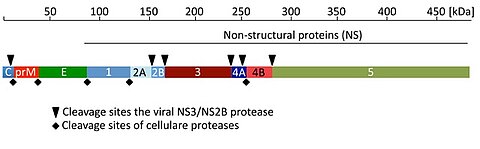
Dengue fever is a severe, widespread and neglected disease with approximately 390 million infections per year. Severe DENV infections and especially reinfections may lead to the dengue hemorrhagic fever and the dengue shock syndrome with lethality up to 5%. The DENV genome contains a single open reading frame, which encodes the structural proteins capsid, membrane precursor (prM), envelope and the non-structural (NS) proteins NS1, NS2, NS3, NS4 and NS5 (Figure 1).
Cellular proteases and the viral serine protease are responsible for cleaving the viral precursor polyprotein into functional proteins (Figure 1). The Dengue virus NS2B/NS3 protease represents a prime target for rational drug design, since protease inhibitors are powerful drugs in Hepatitis C and HIV-1 therapy. However, at the moment there are no clinical PR inhibitors available. We have identified in collaboration with the group of Tanja Schirmeister (University of Mainz) diaryl (thio)ethers as candidates for a novel class of Dengue specific protease inhibitors. These inhibitors block viral replication at submicromolar concentrations and represent a valuable tool for drug development.
Mutations in HIV-1 gag and pol compensate for the loss of viral fitness caused by a highly mutated protease
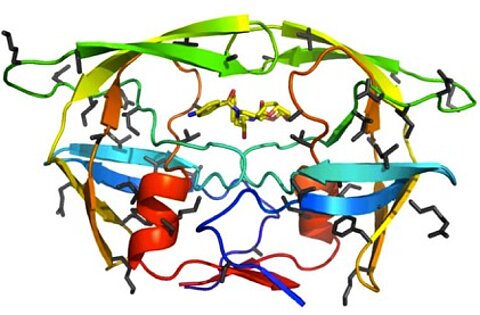
During the last few decades, the treatment of HIV-infected patients by highly active antiretroviral therapy, including protease inhibitors, has become standard. However, development of resistance to antiviral drugs might still occur. Together with Hans-Georg Kräusslich (University of Heidelberg) and with the group of Jan Konvalinka (Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Prague) we have analysed an extreme example of a patient-derived, multi-resistant HIV-1 strain with a highly mutated protease encoding sequence, where up to 19 coding mutations have accumulated in the protease. Biochemical analysis in vitro showed that this protease is highly resistant to most of the currently used protease inhibitors and exhibiting also a very poor catalytic activity. Determination of the crystal structure revealed prominent changes in the protease structure.
While viral loads in the patient were found to be high, insertion of the patient-derived PR into a HIV-1 backbone resulted in reduction of infectivity by three orders of magnitude, but additional introduction of patient derived gag and pol sequences rescued viral infectivity near to wild-type levels. The mutations that accumulated in the vicinity of the processing sites spanning the p2/NC, NC/p1 and p6pol/PR proteins lead much more efficient hydrolysis of corresponding peptides by patient-derived PR in comparison to the wild-type enzyme. This indicates a very efficient co-evolution of enzyme and substrate maintaining high viral loads in vivo under constant drug pressure