Project P01
P01 Integrative analyses of CMV translatomes and MHC-I ligandomes
Prof. Dr. Florian Erhard & Prof. Dr.Lars Dölken
The genomes of cytomegaloviruses (CMV) are among the largest and most complex viral genomes known. Functional genomics methodology including large-scale RNA sequencing (RNA-seq) and ribosome profiling (Ribo-seq) recently resulted in the identification of hundreds of new viral gene products for the human CMV (HCMV) (1, 2). A unified and standardized nomenclature of HCMV gene products is still missing. Accordingly, so far only very few studies have already started to analyze the function of any of the new HCMV gene products. Our data clearly demonstrate a similar picture for the murine CMV (MCMV; Fig.1).
In addition to large viral ORFs of >100 amino acids (aa), cytomegaloviruses encode hundreds of short ORFs (sORFs) of 3 to 99 aa. Many of these represent “upstream open reading frames” (uORFs), which are expressed upstream of annotated larger ORFs in the 5’ leader region of the mRNA previously thought to be untranslated (5’-UTR). The presence of large numbers of viral uORFs is not surprising as more than half of all human genes also express sORFs (3–5)). Interestingly, the vast majority of these do not encode for functional polypeptides as the respective gene products are rapidly degraded upon translation. Recently we were able to deominstrate that sORF-derived polypeptides are nevertheless efficiently presented to patrolling CD8 T cells via MHC-I (2). Priming of T cells against CMV is believed to be mainly mediated by cross-presenting (i.e. non-infected) macrophages. We hypothesize that sORF-derived peptides, which are efficiently presented by MHC-I but of low total abundance in the proteome, represent poor substrates for cross presentation and CD4-CD8 T cell augmentation. These peptides thus represent a new class of antigens. Cellular uORFs orchestrate gene expression by regulating translation initiation of the downstream ORF. We hypothesize that herpesvirus uORFs enable these viruses to adapt viral gene expression to the infected cell type and their local environment to escape detection by the host’s immune system.
In this project, we will provide a state-of-the-art annotation of both the HCMV and MCMV genome. We will utilize this information to identify viral T cell epitopes for novel diagnostic, prophylactic and therapeutic approaches. We will also investigate the immunological importance of viral sORFs in the cellular control of CMV.
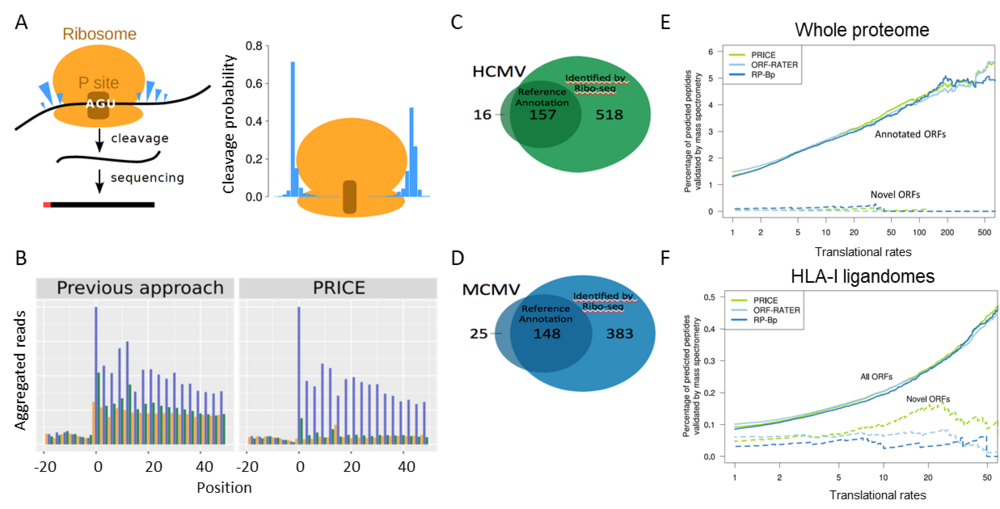
Our computational approach “PRICE” implements a probabilistic model for the stochastic RNase cleavage events in Ribo-seq experiments (A). It thereby substantially reduces experimental noise from Ribo-seq data (B). Global translational activity in HCMV-infected cells relative to the translation start site is depicted (1). Translation in the three different frames is indicated in blue, green and orange. Number of well-expressed viral ORFs for HCMV (C) and MCMV (D). (E) Correlation of translation rates of annotated ORFs and novel sORFs identified in whole proteomes and HLA-I ligandomes using different algorithms (PRICE(6), ORF-RATER (7) or RP-Bp (8)). While sORF-encoded polypeptides remain undetectable by whole proteome mass spec, they are well represented in HLA-I ligandomes.
References
- N. Stern-Ginossar et al., Decoding Human Cytomegalovirus. Science (80-. ). 338, 1088–1093 (2012).
- F. Erhard et al., Improved Ribo-seq enables identification of cryptic translation events. Nat. Methods. 15, 363–366 (2018).
- J. Somers, T. Poyry, A. E. Willis, A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol. 45, 1690–1700 (2013).
- C. Barbosa, I. Peixeiro, L. Romao, Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 9, e1003529 (2013).
- J. I. Pueyo, E. G. Magny, J. P. Couso, New Peptides Under the s(ORF)ace of the Genome. Trends Biochem Sci (2016), doi:10.1016/j.tibs.2016.05.003.
- F. Erhard et al., Improved Ribo-seq enables identification of cryptic translation events. Nat. Methods. 15, 363–366 (2018).
- G. Dunn, J. S. Weissman, Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics. 17, 958 (2016).
- B. Malone et al., Bayesian prediction of RNA translation from ribosome profiling. Nucleic Acids Res (2017), doi:10.1093/nar/gkw1350.